AI-Powered Cell Painting in iPSC-Derived Hepatocytes for Liver Toxicity Profiling
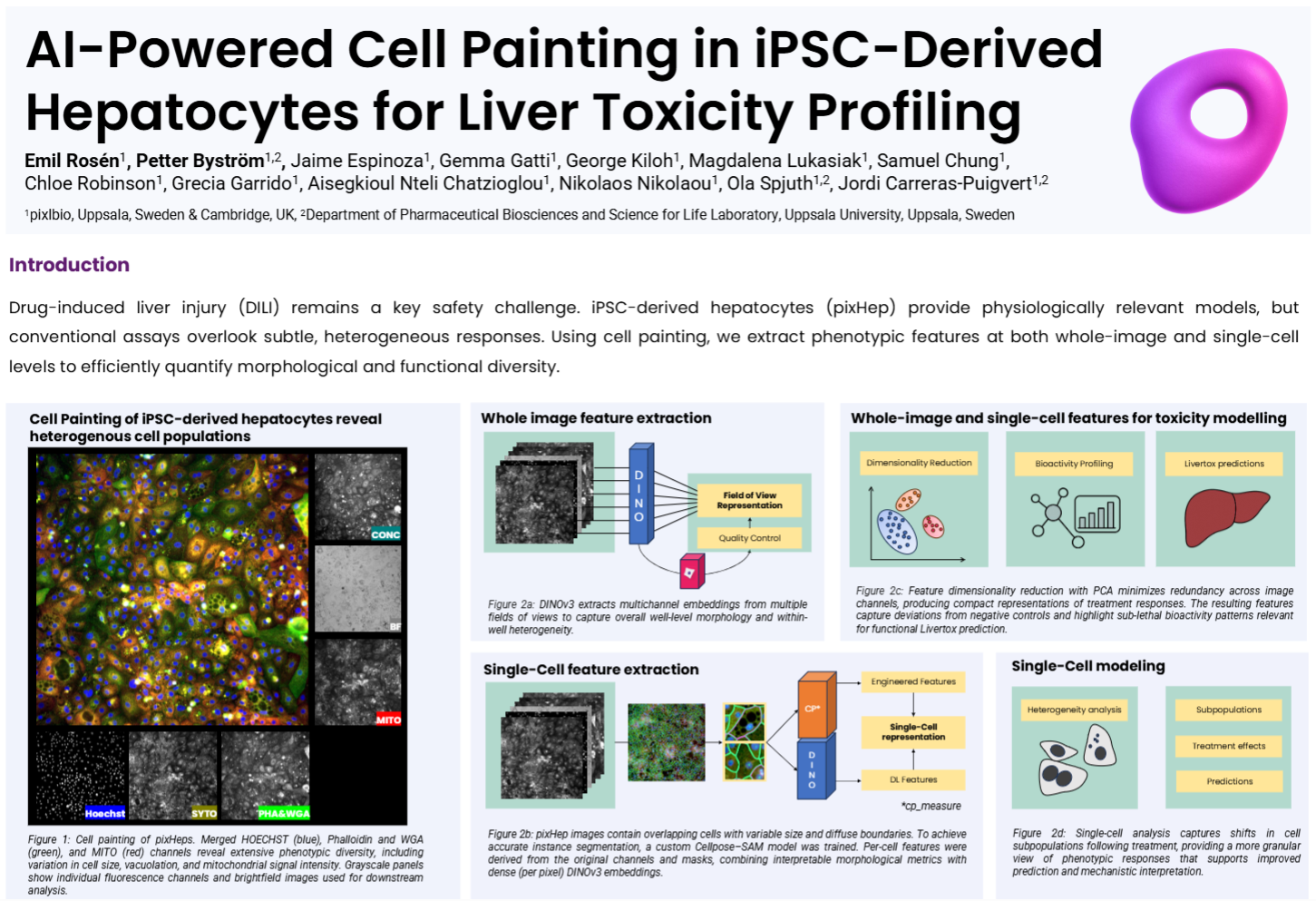

This study applies large-scale Cell Painting to pixlbio’s iPSC-derived hepatocytes (pixHep) to detect early, subtle morphological signatures of drug-induced liver injury across 24 benchmark DILI compounds spanning eight concentrations. Whole-image embeddings generated using DINOv3 captured well-level morphological variance and sublethal bioactivity patterns, while a custom Cellpose–SAM segmentation pipeline enabled per-cell feature extraction that combined interpretable morphology with dense, multichannel embeddings. Both image-level and single-cell representations revealed phenotypic shifts not detectable by viability assays, including compound-specific subpopulations and treatment-driven changes at non-lethal doses. Morphological space analysis showed clear clustering of DMSO controls versus DILI compounds, and models trained on these features achieved up to ~69% balanced accuracy in leave-one-out compound identification and differentiated structurally similar drugs at sub-toxic doses. Single-cell projections further exposed heterogeneous response axes across healthy, stressed, and dying cells, offering finer mechanistic resolution than bulk viability or averaged morphological metrics. Overall, the poster demonstrates that pairing pixHep with modern computer vision models yields a sensitive, scalable foundation for predictive DILI modeling and mechanistic interpretation.



