Catalog Numbers: HEP-003, HEP-059, HEP-004
pixHep iPSC-derived Human Hepatocytes
Human iPSC-derived hepatocytes for predictive disease modeling and toxicity studies.
- Cryopreserved, functionally mature hepatocytes comparable to primary cells
- Proven performance in DILI, MASLD, and metabolism assays
- Consistent, scalable production with 3 available donors off-the-shelf (5 x 10^6 viable cells guaranteed per vial)
pixHep™ are iPSC-derived hepatocytes with mature morphology, functional CYP activity, and stable performance for 30+ days in culture.
They are available in wild-type, disease-specific, or CRISPR-edited formats, enabling precise modelling of metabolism, toxicity, and disease pathways.
Stable Phase I/II enzyme activity for over a month.
Maintains metabolic function for long-term studies, enabling chronic toxicity and metabolite profiling.
ASGR1 is capitalised
Displays native transporter activity for realistic drug absorption and liver-specific distribution studies.
Predictive DILI response matchint clinical outcomes.
Accurately identifies human-relevant drug-induced liver injury during preclinical screening.
Custom genetic variants for mechanistic studies.
CRISPR-engineered pixHeps model disease-linked variants to uncover mechanisms and validate therapeutic targets.
Ready for hign-content imaging and co-culture.
Optimized for complex screening platforms and multi-cell type systems for advanced phenotypic analysis.
Technical Data & Functional Validation
pixHeps demonstrate superiority over conventional IPSC-derived hepatocyte models.
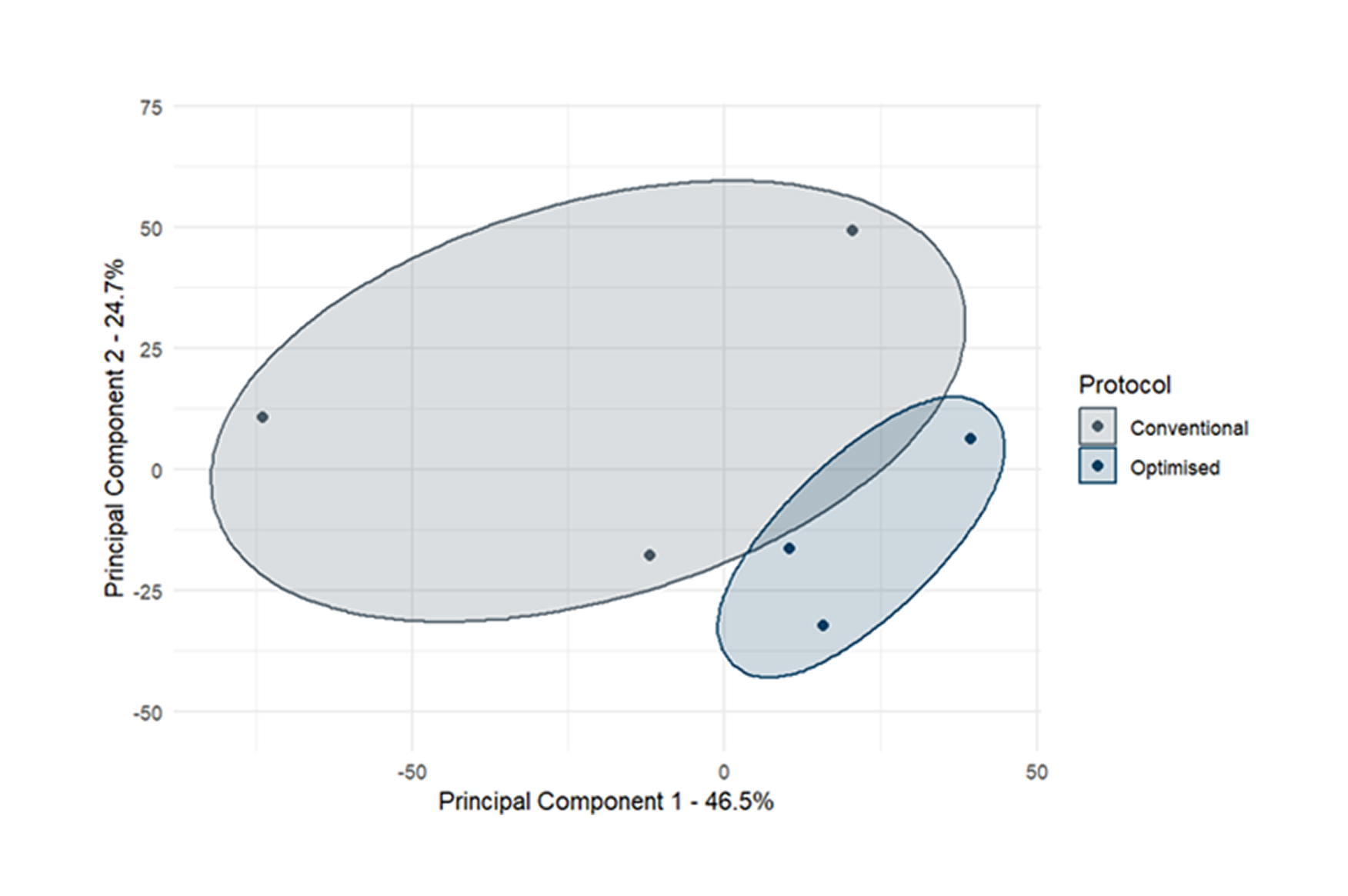

Maturity Markers

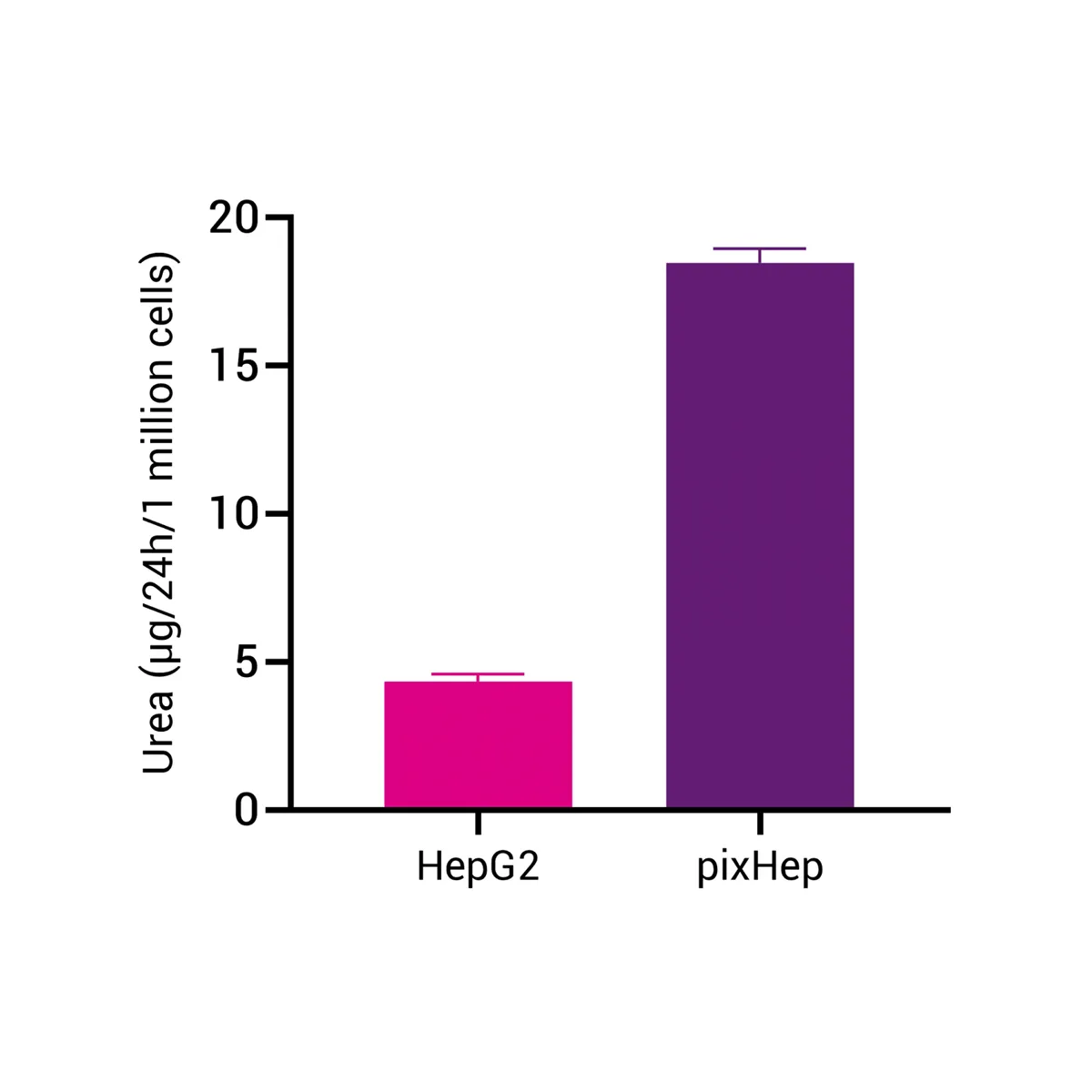
Urea Cycle Markers

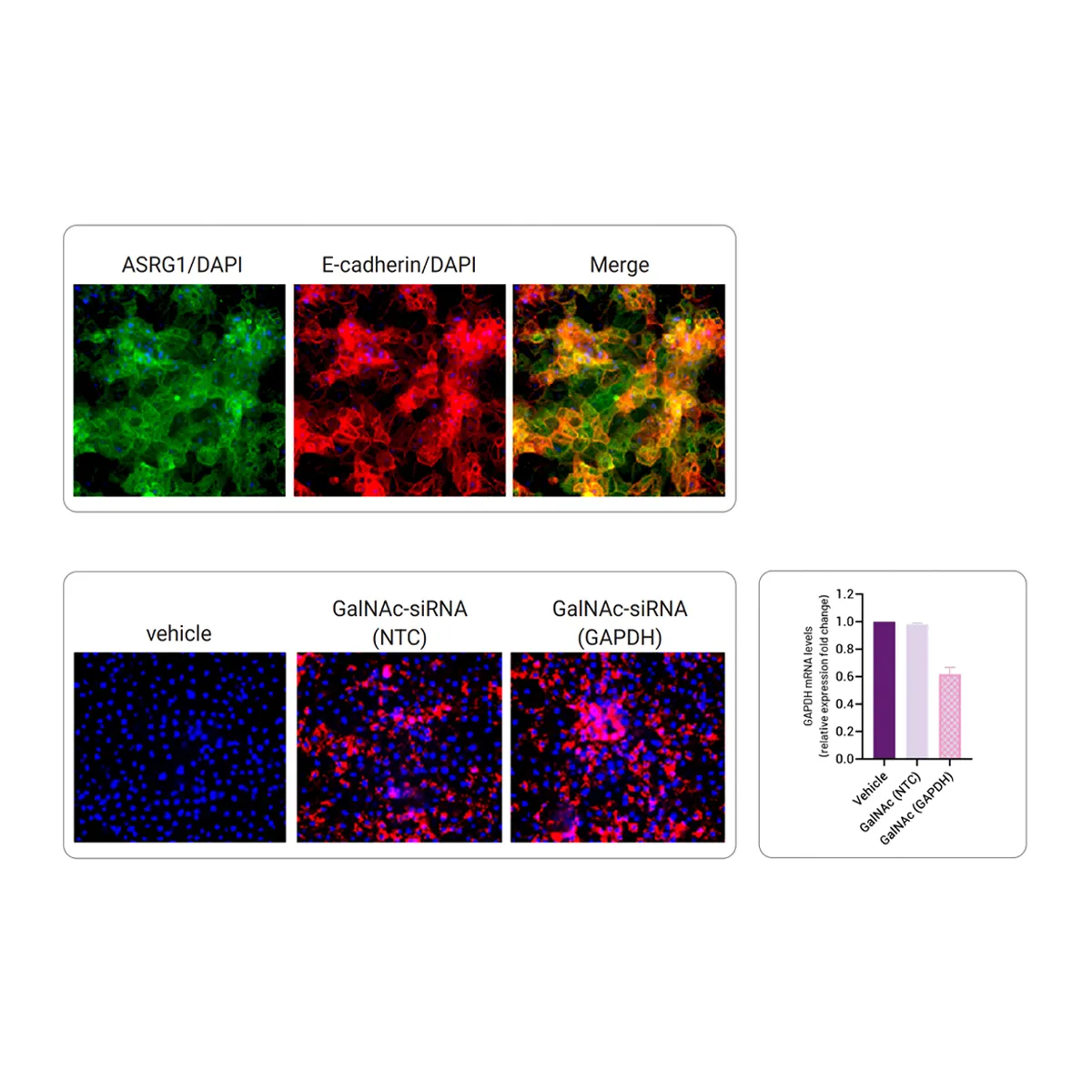
Functional Membrane Localization of ASGR1

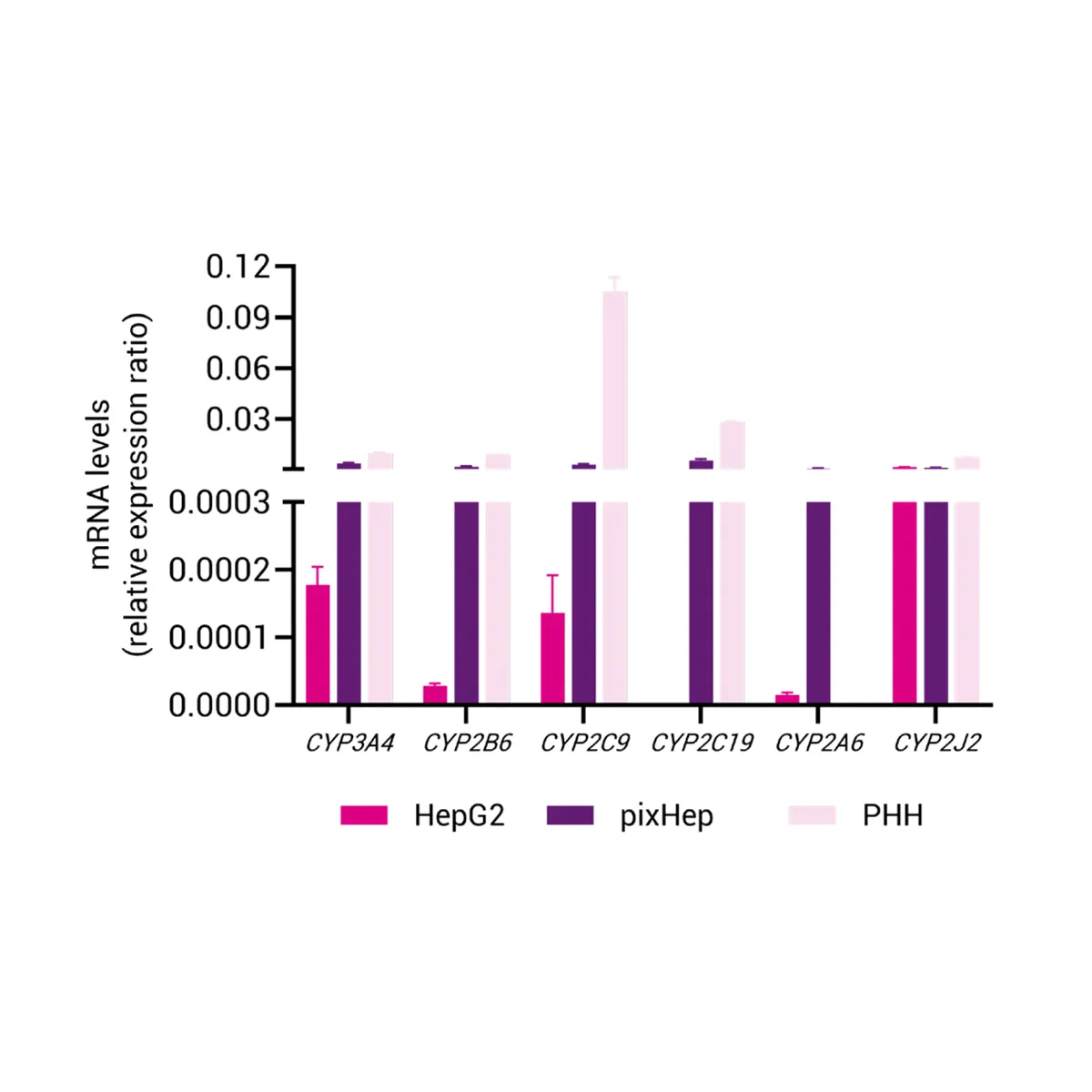
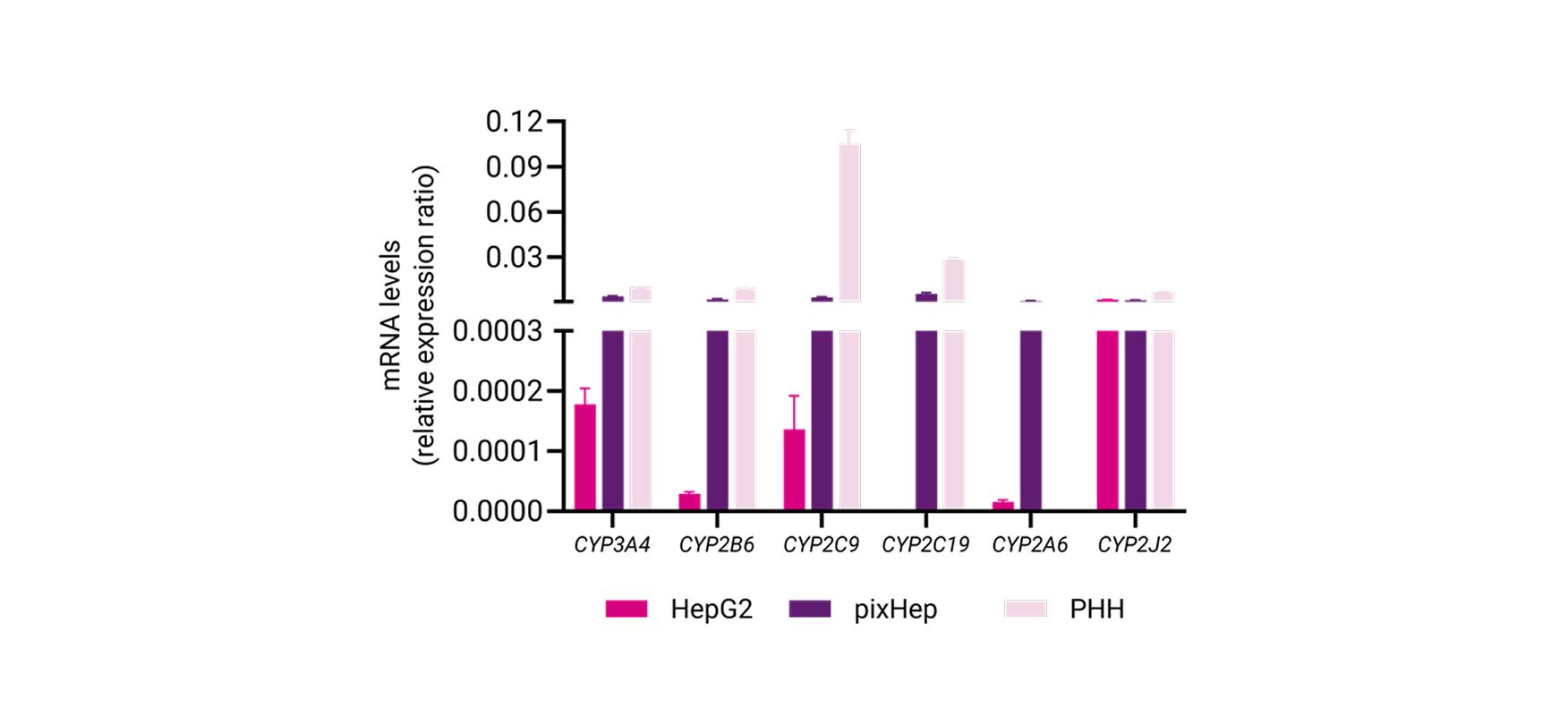
Phase I and Phase II Enzyme Activity
Ready to Turn Your Cells Into Data?