Catalog Numbers: HEP-003, HEP-059, HEP-004, HEP-003-TM6SF2-a, HEP-003-PNPLA3-h
MASLD (Metabolic Dysfunction-Associated Steatotic Liver Disease) Models
iPSC-derived hepatocyte models replicating steatosis, insulin resistance, and inflammation.
- Cryopreserved vials (1 × 10⁶ viable cells per vial) from defined donor backgrounds, PNPLA3, TM6SF2, and multiple wild-type backgrounds available
- Inducible steatosis and lipid accumulation validated by BODIPY and cytokine readouts
- Suitable for mechanistic and compound screening in metabolic disease pathways
Place your Order
Our MASLD pixHep™ model replicates the lipid accumulation, metabolic dysfunction, and inflammatory signalling of clinical steatosis.
Available in wild-type or genetic risk backgrounds, it provides a robust platform for screening metabolic modulators, anti-inflammatories, and disease progression blockers.
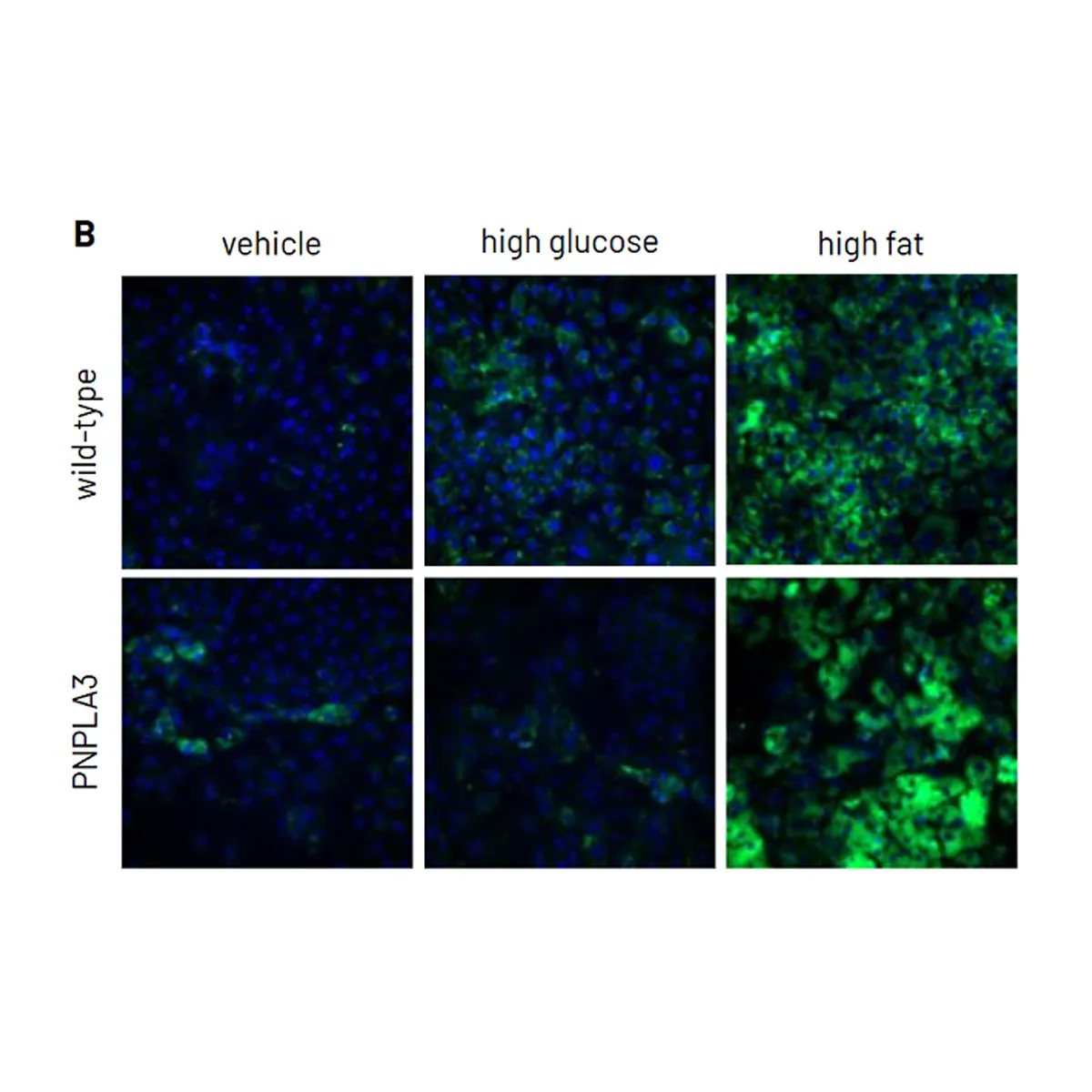
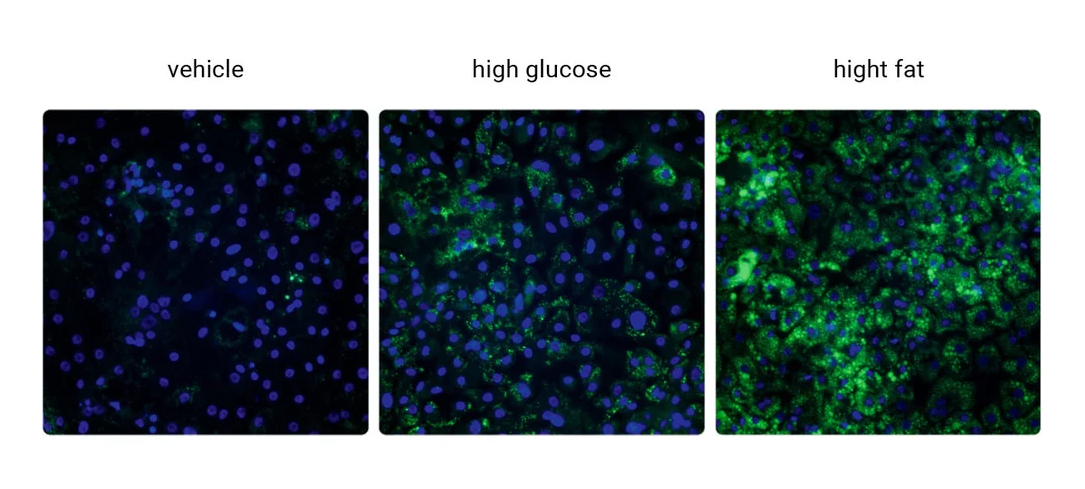
Lipid droplet accumulation confirmed by imaging.
Altered lipid metabolism and oxidative stress markers.
Compatible with CRISPR-driven genetic risk variants.
Co-culture ready for MASH progression modelling.
Technical Data & Functional Validation
Increased Lipid Accumulation

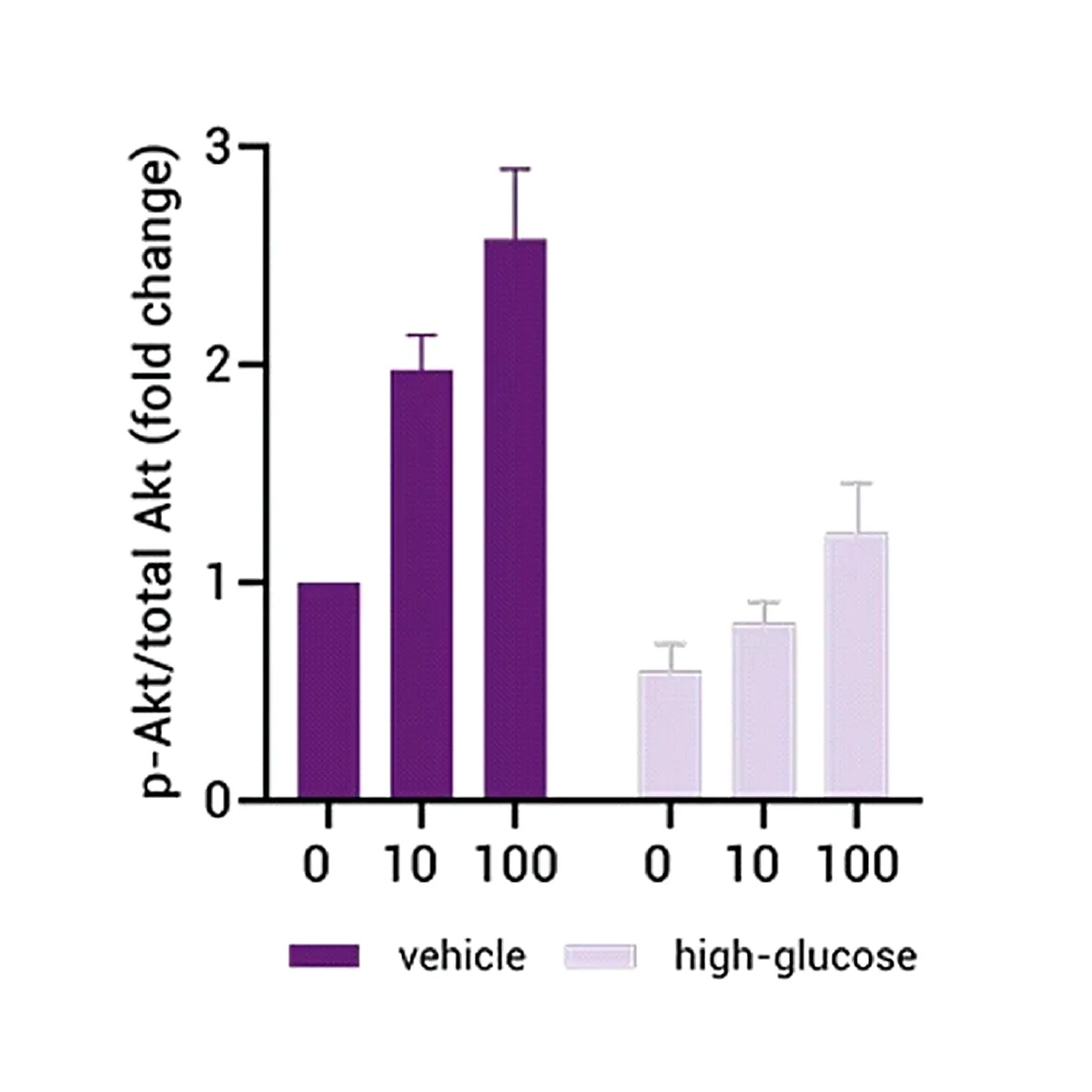
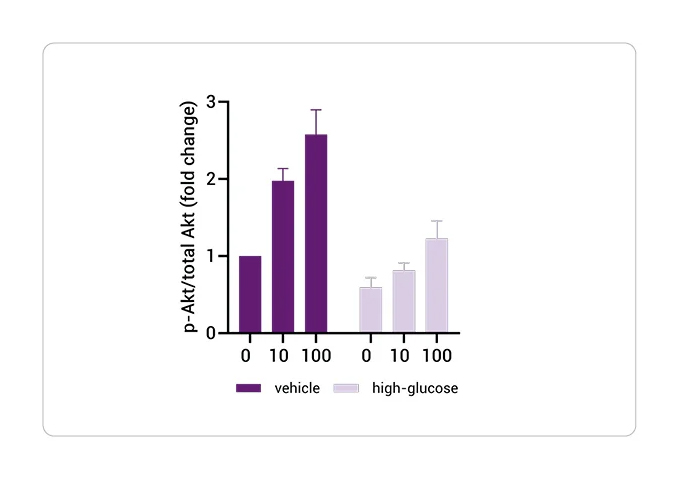
Insulin Resistance
Lipotoxicity and Pro-Inflammation

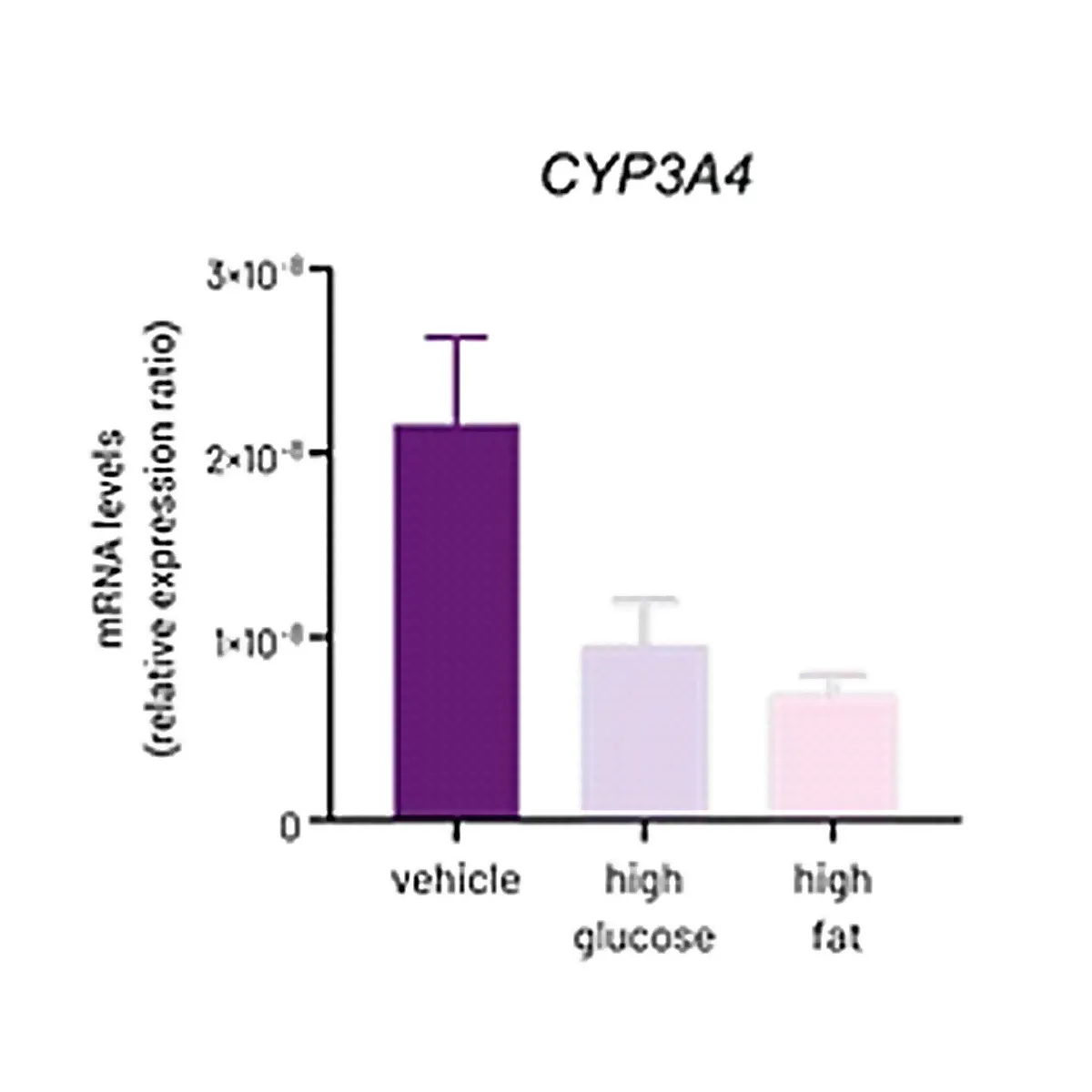
Reduced Phase 1 Activity

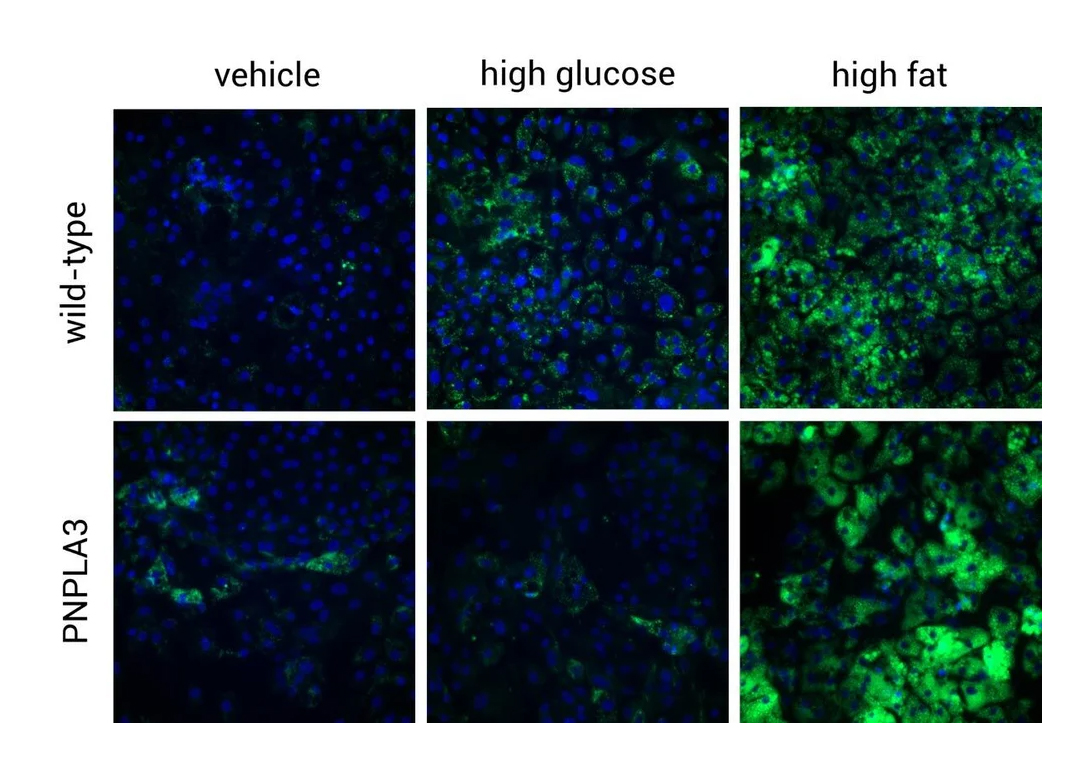
PNPLA3 Variant MASLD Phenotype
Streamline Your Research
Ready to Turn Your Cells Into Data?
Our pixCell portfolio seamlessly integrates into custom data generation projects—from functional assays (pixCellServices) to phenomic analysis (pixCellPaint)—delivering ready-to-use data at your fingertips.